
This is a repository for the associated Python code for the manuscript titled: From spikes to intercellular waves: tuning intercellular Ca2+ signaling dynamics modulates organ size control. The code was built by Dr. Ramezan Paravi Torghabeh. The code was then later updated and adapted for creation of the figures in the manuscript by Francisco Huizar. Experimental work and validation was led by Dharsan Soundarrajan. This work was done within the Multicellular Systems Engineering Lab at the University of Notre Dame. Please direct any questions to the lab principal investigator, Dr. Jeremiah Zartman.
All code was done using Jupyter notebooks where all code was run and the visible outputs are saved in the iPython notebooks. The code is available to be downloaded and run after installation of the latest version of Jupyter and Python. All simulation outputs were coded to be saved in the simulationResults folder, and are labeled according to figure numbers. Use of Epitools was done to create a 2-D geometrically accurate representation of the Drosophila wing imaginal disc. The corresponding folder for this is in the epitools (Figure S1 Creation) folder here.
Sample demonstration of the code
Anyone can run a sample of the code used in the manuscript by using the Google Colaboratory notebook located here. Instructions to run the demonstration code are found on the linked page. A full demonstration simulation will take approximately 25 minutes to run on the Google Colaboratory virtual machine, and simulations can be run faster on a personal machine (see instructions below for use on a personal machine). In the demonstration page, you are able to choose between the following outputs that correspond to Figure 2 in the manuscript:
- Single cell calcium spikes
- Intercellular calcium transients
- Intercellular calcium waves
- Global fluttering calcium activity
Open-source license agreement
- This code is made publicly available for use under the GNU Lesser General Public License, version 2.1
- You can find the license agreement for this code here
- We kindly ask users of the code and any derivative work to cite the original publication:
Soundarrajan, D. K., Huizar, F. J., Paravitorghabeh, R., Robinett, T. & Zartman, J. J. From spikes to intercellular waves: Tuning intercellular calcium signaling dynamics modulates organ size control. PLOS Computational Biology 17, e1009543 (2021).
The below licensing statements are verbatim statements from the Free and Open Source Software Auditing (FOSSA) team originating from this webpage
Users of this code must:
- Include a copy of the full license text and the original copyright notice
- Make available the source code when you distribute a derivative work based on the licensed library
- License any derivative works of the library under the same or later version of the LGPL or GPL
The LGPL license allows users of the licensed code to:
- Use the code commercially: Like GPL, LGPL imposes no conditions on using the code in software that’s sold commercially
- Change the code: Users can rework the code, but if they distribute these modifications, they must release these updates in source code form
- Place warranty: Distributors of the original code can offer their own warranty on the licensed software
Instructions to run the code on a personal machine
- You can download all of the associated code here
- Each notebook is labeled corresponding to its associated figure in the main text of the manuscript.
- NOTE: The full folder is approximately 2+ GB of data as it contains the pre-run simulation output pictures and videos.
- Extract the downloaded .zip folder where you would like to run the code using Jupyter

- Navigate to and open the extracted folder
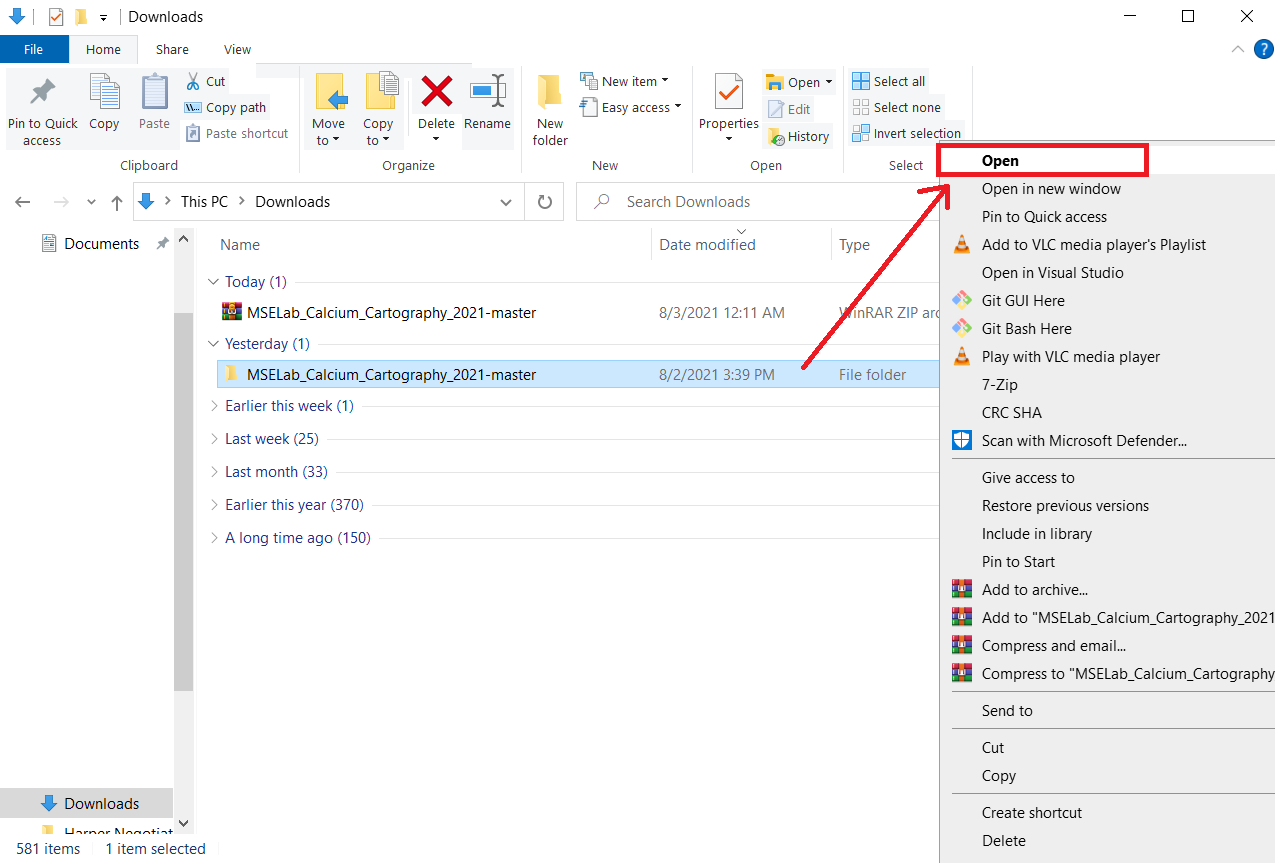
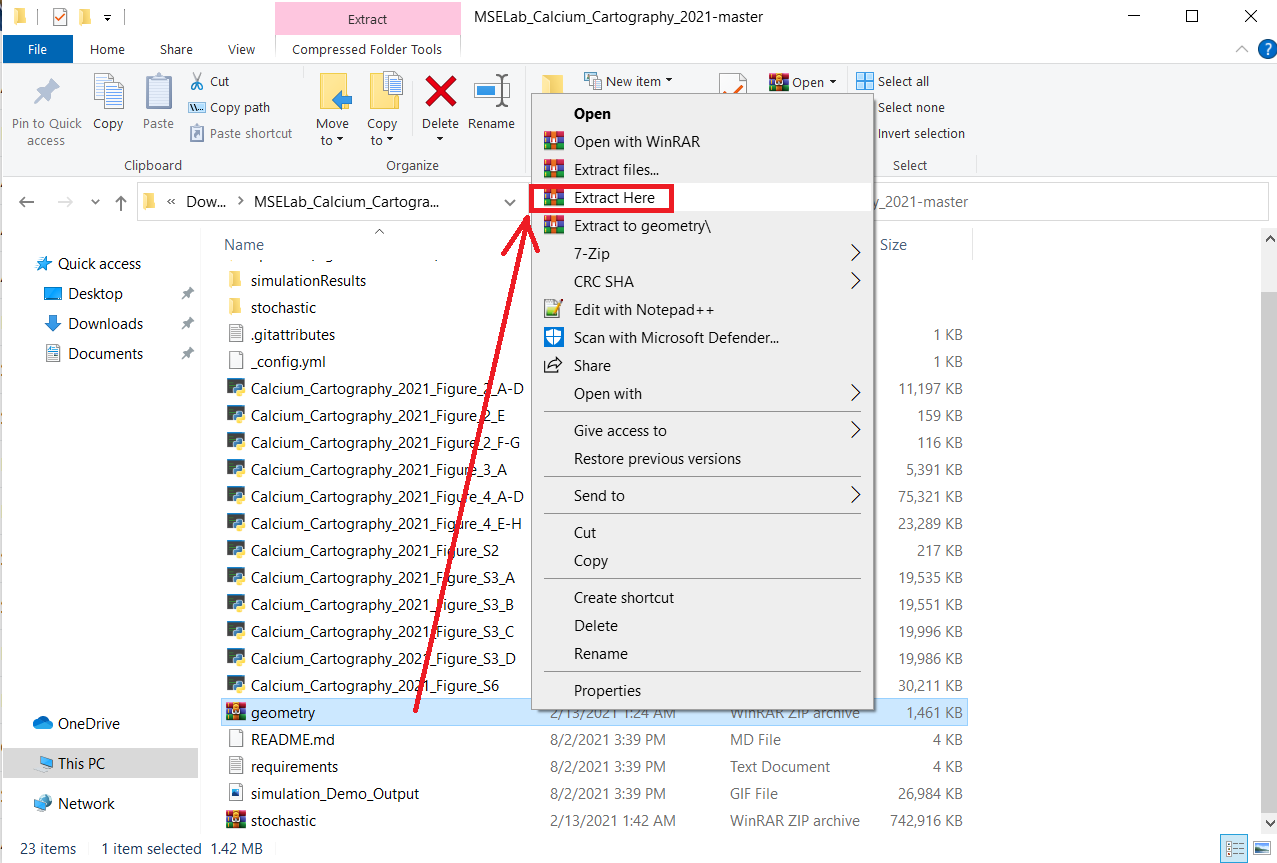
- Within the now extracted folder extract the ‘geometry.zip’ folder as well
- Open and run Jupyter Notebook
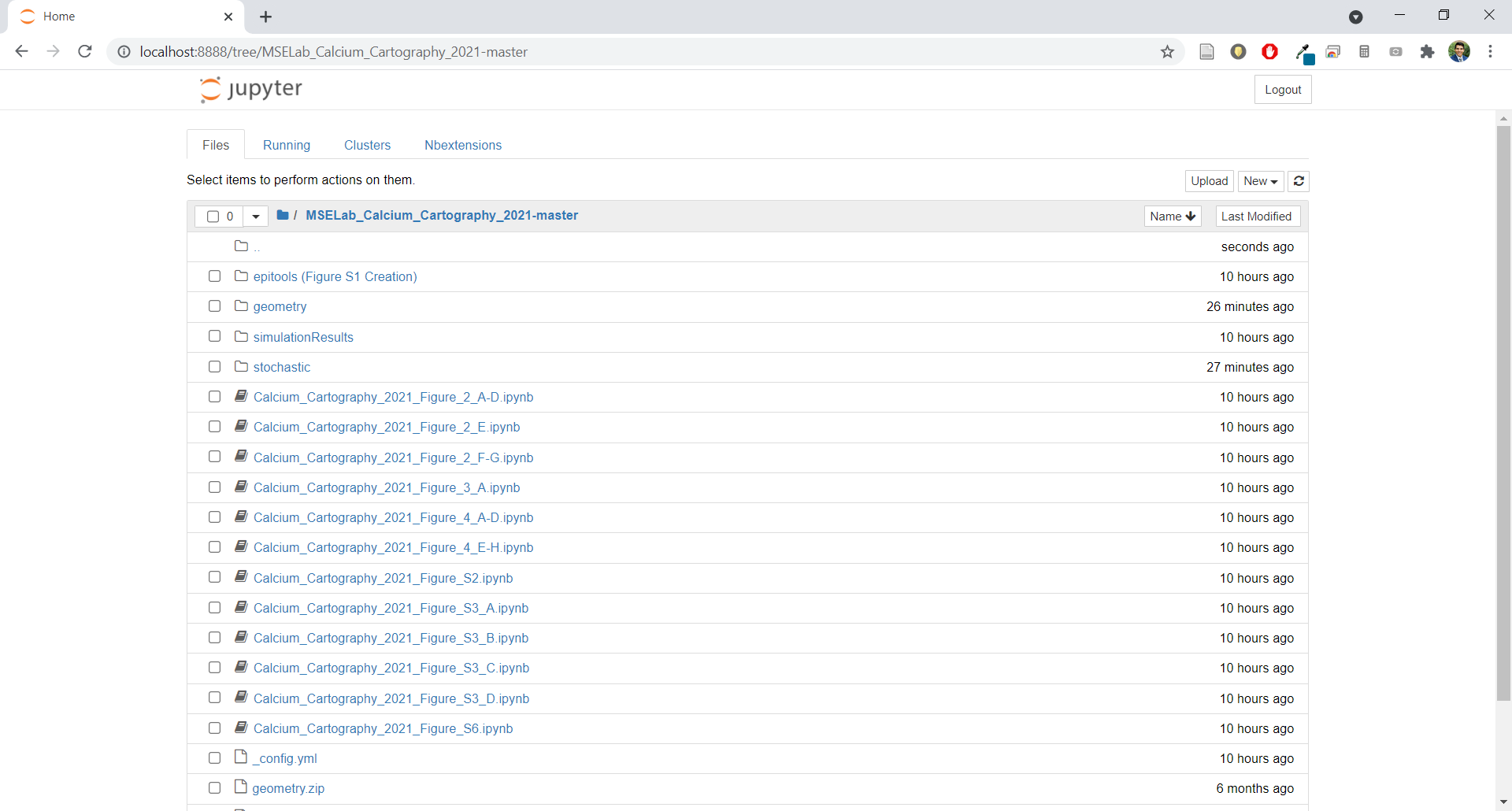
- Navigate in the Jupyter Notebook host to where the extracted files are
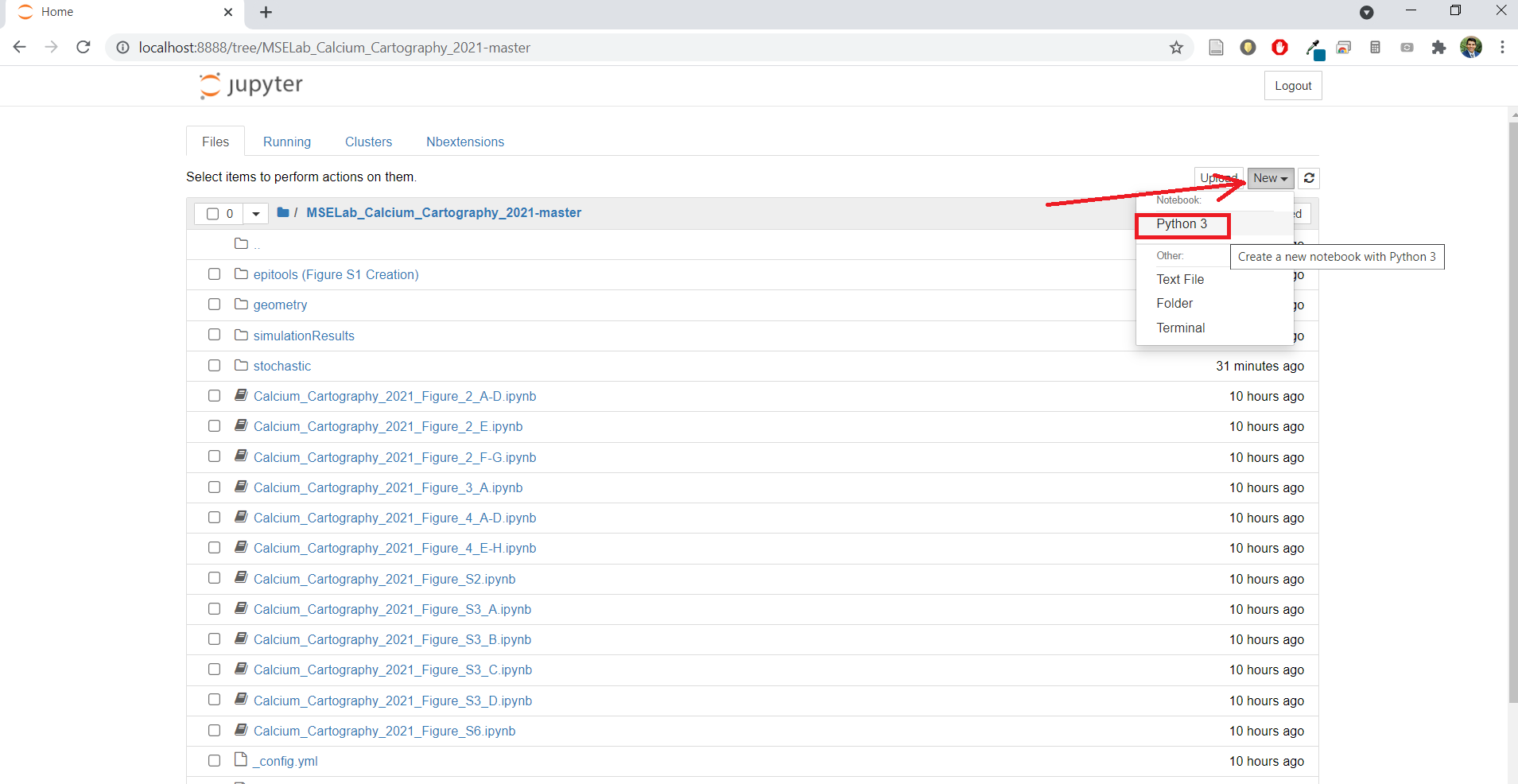
- Open a new, untitled notebook in Jupyter
- Install all necessary Python packages using pip install requirements.txt.
- This is done by entering the following into a cell:
!pip install -r /path/to/requirements.txt - NOTE: ‘path/to/’ should be changed to the correct path where the code was extracted to
- This is done by entering the following into a cell:
- Use Jupyter to open each individual notebook to run the simulations as desired
Code for repeated simulations
You can find the code and corresponding outputs for repeated simulations (with different random number generator seeds) for the manuscript’s main figures in the following locations:
Code for model sensitivity to parameters
You can find the code and corresponding outputs for the sensitivity analysis of parameters and their effects on a baseline intercellular calcium wave here. This code corresponds to the supplementary information figures.
Acknowledgements
We would like to thank Trent Robinett for his contributions towards completion of the manuscript. In particular, we would like to highlight his efforts in carrying out experiments alongside Dharsan Soundarrajan.
The work in this manuscript was supported by NIH Grant R35GM124935 and NSF Award CBET-1553826. The authors gratefully acknowledge the Notre Dame Center for Research Computing (CRC) for providing computational facilities in addition to members of the Zartman Lab for helpful discussions and feedback.